2021-03-25
Back to listCompound screening to find molecules with the desired biological activity is of great importance to drug design. Traditional screening methods require synthesis and the biological testing of large compound collections, which can turn into a costly and time-consuming process with a low success rate. Machine learning, particularly graph neural networks, has the potential to replace traditional methods by enabling AI assisted virtual screening. This will further speed intermediate steps and significantly reduce R&D costs.
We are excited to announce that our AI models, trained with Baidu’s open-source deep learning platform PaddlePaddle, ranked first on a well-recognized benchmark leaderboard for molecular property predictions. All algorithms and pretrained models stem from Paddle Graph Learning (PGL), a graph learning framework, and PaddleHelix, a machine learning bio-computing framework.
The HIV and PCBA datasets from Open Graph Benchmark (OGB), a set of benchmark datasets aiming to facilitate graph learning research, are among the world’s largest benchmarks for molecular property prediction. The HIV task is to predict whether a compound inhibits HIV virus replication or not. The PCBA task is to classify compounds based on their effectiveness against over 100 disease targets. For example, compounds that can increase expression of functional SMN2 protein can alleviate spinal muscle atrophy caused by a mutation of SMN1 protein.
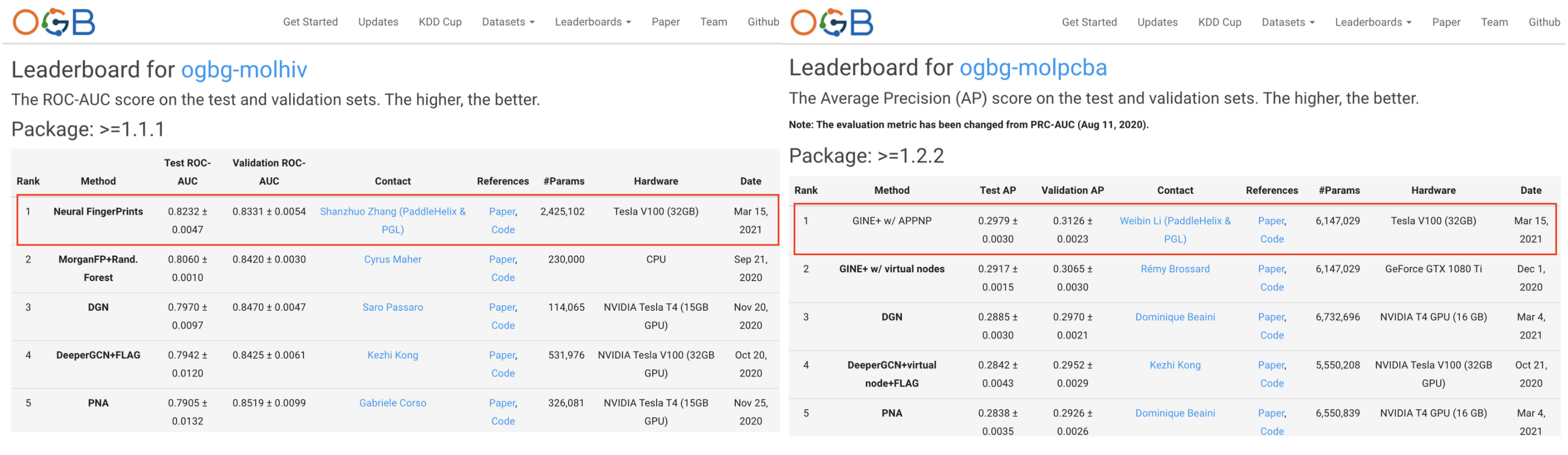
To tackle the challenge, our researchers trained a deep graph neural network to learn molecular chemical representation through a self-supervised learning task. Additionally, they took the representation to train a molecule property prediction classifier.
Molecular representation learning
In the HIV task, the first step is to learn the molecule chemical representation with the graph neural network. OGB provides graph representations of molecules where nodes are atoms, and edges are chemical bonds alongside atom features. However, we found that these features could not represent the chemical information of molecules without considering domain knowledge.
Our researchers trained a graph neural network to learn molecular representation by integrating its chemical properties, such as fingerprints, the presence of absence of particular chemical substructures. This novel representation learning method plays a critical role in achieving the SOTA results on the OGB leaderboard for HIV.
Graph learning technology
In the PCBA task, our researchers managed to improve the performance by integrating GINE plus and the APPNP algorithm based on PGL without adding extra model parameters. APPNP algorithm can utilize the relationship between graph convolutional networks (GCN) and PageRank to derive an improved propagation scheme based on personalized PageRank.
What is PGL and PaddleHelix?
PGL is an efficient and flexible graph learning framework based on PaddlePaddle, which was open sourced in 2019. Its latest version 2.0 supports dynamic (computational) graphs and large-scale graphs. Developers can use PGL to efficiently build graph neural networks for industrial applications, ranging from recommendation systems to search engines, finance, maps, security risk control, and biomedicine.
PaddleHelix is a machine learning bio-computing framework built upon PaddlePaddle, aiming to facilitate the development of vaccine design, drug discovery, and precision medicine.Open sourced in December, 2020, PaddleHelix currently provides pretrained models including representation learning for compounds and proteins, LinearRNA, drug-target interaction, and ADMET modeling. With PaddleHelix, we are aiming to provide an industry-facing bio-computing ecosystem and services in the future.