2023-05-03
Back to list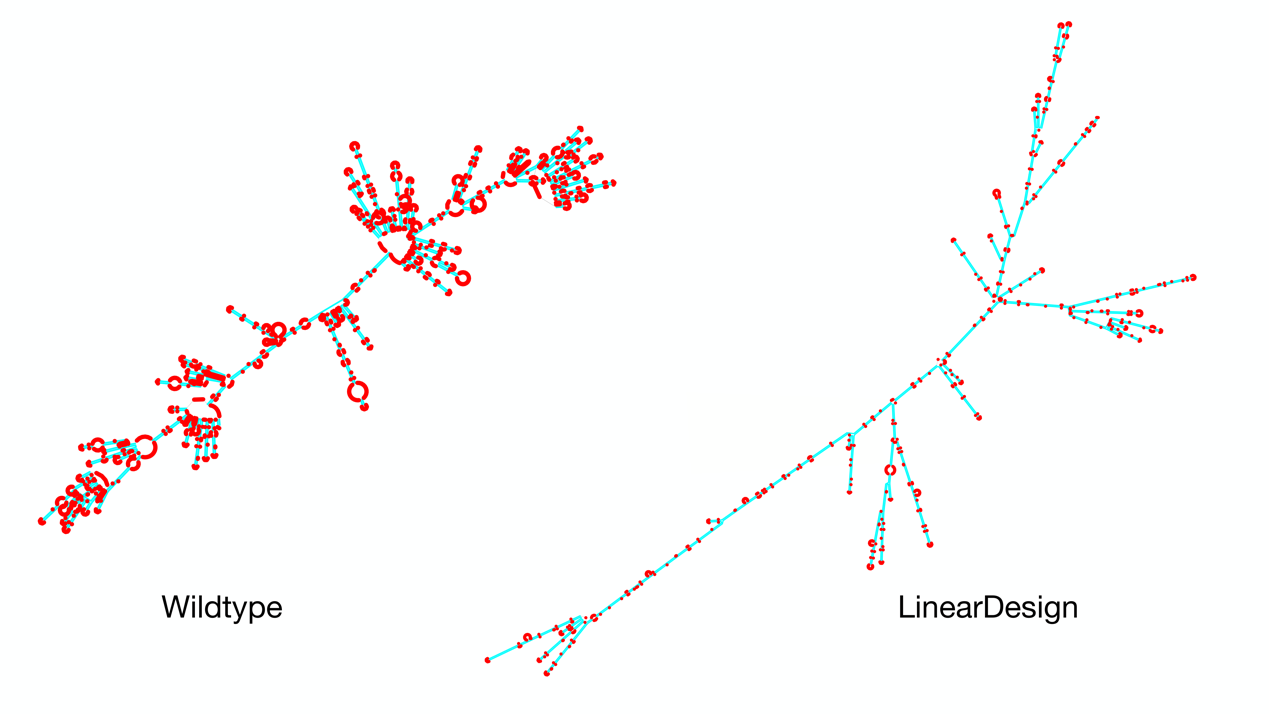
- The study “Algorithm for Optimized mRNA Design Improves Stability and Immunogenicity”, featuring Baidu Research as the first affiliation, appeared in the scientific journal Nature today.
- LinearDesign achieves an impressive 128-fold increase in the COVID-19 vaccine's antibody response. Its applications extend beyond vaccines to encompass all therapeutic proteins.
- The paper reveals how a complex biology problem can be tackled by taking an elegantly simple technique from natural language processing (NLP).
A team of researchers from Baidu Research has developed an AI algorithm that can rapidly design highly stable COVID-19 mRNA vaccine sequences that were previously unattainable. The algorithm, named LinearDesign, represents a major leap in both stability and efficacy for vaccine sequences, achieving a 128-fold increase in the COVID-19 vaccine’s antibody response.
“This research can apply mRNA medicine encoding to a wider range of therapeutic proteins, such as monoclonal antibodies and anti-cancer drugs, promising broad applications and far-reaching impact,” said Dr. He Zhang, Staff Software Engineer at Baidu Research.
Through a collaboration with Oregon State University, StemiRNA Therapeutics, and the University of Rochester Medical Center, the study “Algorithm for Optimized mRNA Design Improves Stability and Immunogenicity” appeared in the scientific journal Nature today through Accelerated Article Preview (AAP). This marks the first time a Chinese tech company has been credited as the first affiliation on a paper published in Nature. The paper reveals how a complex biology problem can be tackled by taking a classic approach from natural language processing (NLP), using an elegantly simple solution that has been employed to understand words and grammar.
mRNA, or Messager RNA, has emerged as a revolutionary technology for vaccine development and potential treatments against cancer and other diseases. Serving as a vital messenger that carries genetic instructions from DNA to the cell’s protein-making machinery, mRNA enables the creation of specific proteins for various functions in the human body. With numerous advantages in safety, efficacy, and production, mRNA has been swiftly adopted in the process of COVID-19 vaccine development.
However, the natural instability of mRNA results in insufficient protein expression that weakens a vaccine’s capacity to stimulate strong immune responses. This instability also poses challenges for storing and transporting mRNA vaccines, especially in developing countries where resources are often limited.
Previous research has shown that optimizing the secondary structure stability of mRNA, when combined with optimal codons, leads to improved protein expression. The challenge lies in the mRNA design space, which is incredibly vast due to synonymous codons. For instance, there are approximately 10^632 mRNAs that can be translated into the same SARS-CoV-2 Spike protein, presenting insurmountable challenges for prior methods.
Though NLP and biology may at first glance appear unrelated, the two fields share strong mathematical connections. In human language, a sentence consists of a word sequence and an underlying syntactic tree with noun and verb phrases, which together convey meaning. Likewise, an RNA strand has a nucleotide sequence and an associated secondary structure based on its folding pattern.
Researchers used a technique in language processing called lattice parsing, which represents potential word connections in a lattice graph and selects the most plausible option based on grammar. Similarly, they created a graph that compactly represents all mRNA candidates, using deterministic finite-state automaton (DFA). Applying lattice parsing to mRNA, finding the optimal mRNA is akin to identifying the most likely sentence among a range of similar-sounding alternatives.
Using this approach, LinearDesign takes a mere 11 minutes to generate the most stable mRNA sequence that encodes Spike protein.
In a head-to-head comparison, the sequences designed by LinearDesign exhibited significantly improved results compared to existing vaccine sequences. For COVID-19 mRNA vaccine sequences, the algorithm achieved up to a 5-fold increase in stability (mRNA half-life), a 3-fold increase in protein expression levels (within 48 hours), and an incredible 128-fold increase in antibody response. For VZV mRNA vaccine sequences, the study reported up to a 6-fold increase in stability (mRNA molecule half-life), a 5.3-fold increase in protein expression levels (48 hours), and an 8-fold increase in antibody response.
“The vaccines designed through our method may offer better protection with the same dosage, and potentially provide equal protection with a smaller dose, leading to fewer side effects. This will greatly reduce the vaccine research and development costs for biopharmaceutical companies while improving the outcomes,” Dr. Zhang added. In 2021, Baidu and Sanofi began a partnership to integrate the LinearDesign algorithm into Sanofi's product design pipeline for mRNA vaccine and drug development.
Baidu has created a biocomputing platform based on PaddlePaddle called PaddleHelix, which encompasses the ERNIE-Biocomputing Big Models, including LinearDesign. This platform explores the application of AI in various fields, such as small molecules, proteins/peptides, and RNA, offering a novel research paradigm for AI in life sciences. Baidu’s ERNIE Big Models have developed a comprehensive big model technology system, covering NLP, vision, cross-modal, and biocomputing. The recently unveiled ERNIE Bot, a knowledge-enhanced large language model (LLM) capable of understanding and generating human language, is part of the ERNIE Big Model family.
Moving forward, Baidu will continue to explore AI applications in life sciences, broadening the scope and depth of inclusive technology, and championing the health and well-being of all humanity.
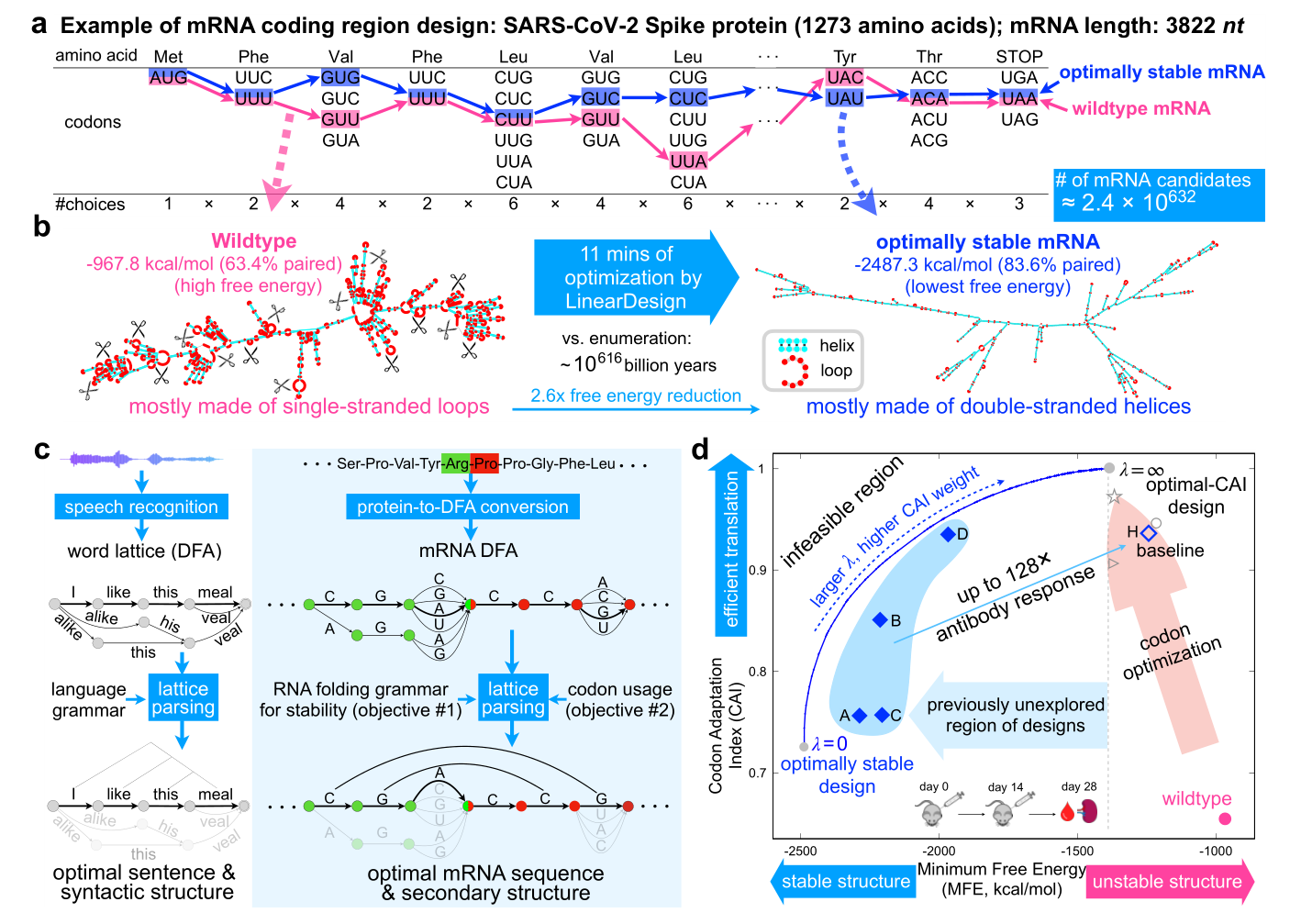
Fig. 1 | Overview of mRNA coding region design for two well-established objectives, stability and codon optimality, using SARS-CoV-2 Spike protein as an example. a, The combinatorial nature of mRNA design due to codon degeneracy (~10632 mRNA sequences for the Spike protein; taking ~10616 billion years to enumerate). The pink and blue paths represent the wildtype and the optimally stable (i.e., lowest energy) sequences, respectively. b, The vastly different secondary structures between these two sequences, with the former being mostly single-stranded (prone to degradation in red loop regions) while the latter mostly double-stranded. Our algorithm takes just 11 minutes for this optimization. c, An analogy between linguistics (left) and biology (right), where deterministic finite-state automaton (DFA) and lattice parsing from the former were adapted to solve mRNA design. An mRNA DFA (inspired by “word lattice”) compactly encodes all mRNA candidates, which are folded simultaneously by lattice parsing to find the optimal mRNA (Fig. 2). d, 2D visualization of the mRNA design space, with stability on the x-axis and codon optimality on the y-axis. The standard mRNA design method codon optimization improves codon usage (the pink arrow) but is unable to explore the vast high-stability region (left of the dashed line), which is exemplified by the COVID-19 vaccine products of BioNTech-Pfizer (○), Moderna (☆), and CureVac (▷). LinearDesign, by contrast, jointly optimizes stability and codon optimality (the blue curve, with λ being the weight of the latter). By considering other factors, we select seven of our designs (four shown here) for COVID-19 vaccine experiments (Fig. 4), which show substantially enhanced half-life and protein expression, and up to 128 antibody responses over the codon-optimized baseline (H). Experiments on the varicella-zoster virus (VZV) mRNA vaccine (on a different antigen, and with different UTRs) show similar improvements (Fig. 5), confirming the generalizability of LinearDesign.
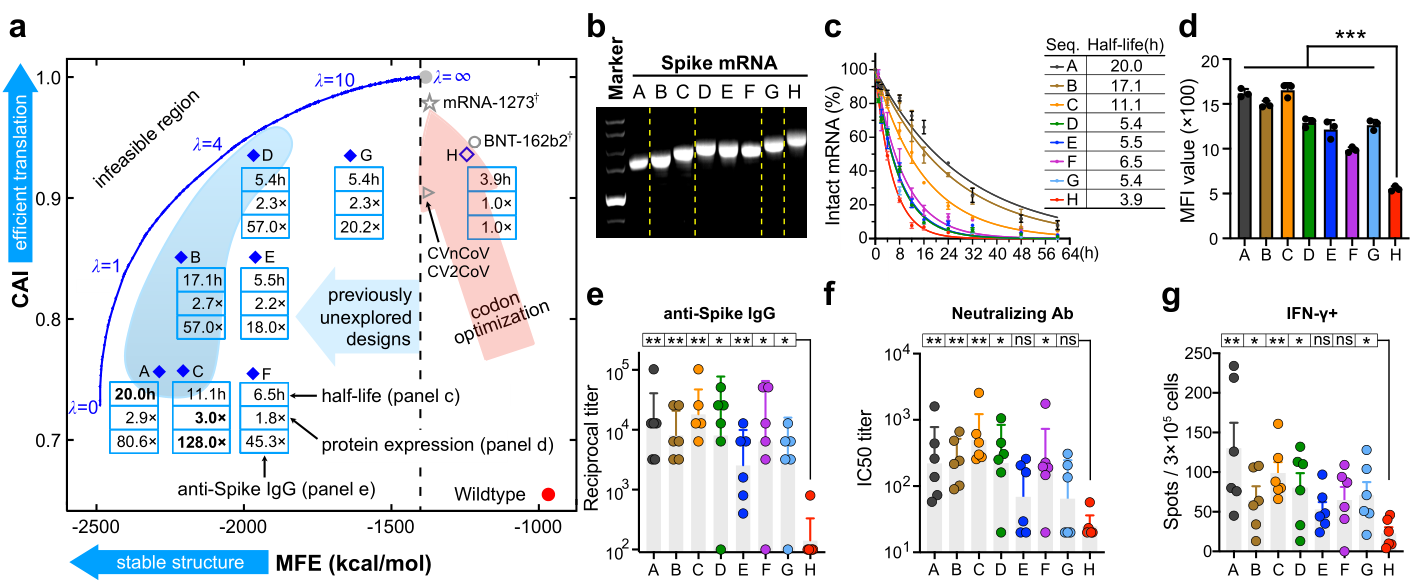
Fig. 4 | Experimental evaluation of LinearDesign-generated mRNA sequences encoding SARS-CoV-2 Spike protein. a, Summary of chemical stability, protein expression of our mRNA designs (A–G) and their immunogenicity in the induction of anti-Spike IgG compared to the codon-optimized baseline (H). b, Non-denaturing agarose gel characterization of mRNA showing the correlation of gel mobility with minimum free energy; for gel source data, see Supplementary Fig. 1a. c, Chemical stability of mRNAs upon incubation in buffer (Mg2+ = 10 mM) at 37 °C. Percentage of intact mRNA is shown. Data is from three independent experiments. d, Protein expression levels of mRNAs determined by flow cytometry 48 hours after transfection into HEK-293 cells. Mean fluorescence intensity (MFI) values derived from three independent experiments are shown. Kruskal–Wallis analysis of variance (ANOVA) with Dunn’s multiple comparisons test to H group was performed for statistical analysis. e–g, C57BL/6 mice (n=6) were immunized i.m. with two doses of formulated mRNA at a 2-week interval. Endpoint titer of anti-Spike IgG (e). Levels of neutralizing Abs against wide-type SARS-CoV-2 (f). Frequencies of IFN-γ-secreting T cells measured by ELISpot (g). A two-tailed Mann-Whitney U test was used for statistical analysis. *p < 0.05, **p < 0.01, ***p < 0.001. Data are presented as mean ± s.d. (c, d), geometric mean ± geometric s.d. (e, f) or mean ± s.e.m. (g). See Source Data for details. See also Extended Data Figs. 5–7, Supplementary Figs. 10 and 12, and Supplementary Tab. 2.