2018-06-18
Back to listBy Yi Li, Wei Ping
While there are various methods to diagnose cancer, a pathology review of biopsied tissues is often considered the gold standard. However, reviewing pathology slides is not easy, even for experienced pathologists. A digitalized pathology slide at 40X magnification often contains billions of pixels, and may take up multiple gigabytes of disk space. In these massive files, pathologists sometimes have to look for micrometastases, small groups of tumor cells that are an early sign for cancer. These tumor cell groups can be smaller than 1000 pixels in diameter1. This makes reviewing pathology slides without missing any of these tiny but clinically actionable evidences very complex and time consuming. Figure 1 illustrates this difficult task.

Various deep learning based algorithms have been proposed to aid pathologists in effectively reviewing these slides and detecting cancer metastasis. Because of the outrageously large size of the original digital slides, most of the algorithms currently being used split the slide into lots of smaller individual image patches, e.g. 256x256 pixels. A deep convolutional neural network is then trained to classify whether each small patch contains tumor cells or normal cells separately. However, sometimes it is difficult to predict whether a patch contains tumor cells without knowing its surroundings, especially around the tumor/normal boundary regions, and false positive predictions are often introduced. Figure 2 shows one example of how difficult this can be.

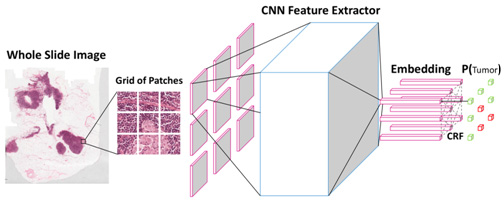
We have proposed a new deep learning algorithm that takes not just one individual patch but a grid of neighboring patches as input to jointly predict whether they are tumor cells or normal cells. This technique could be compared to a pathologist zooming out to see the larger field and make more confident judgements. The spatial correlations between neighboring patches are modelled through a specific type of probabilistic graphical model named conditional random fields. The whole deep learning framework can be trained end-to-end on GPU without any post processing. Figure 3 shows the architecture of the algorithm.
By considering the spatial correlations between neighboring patches, our algorithm introduced far fewer false positives. Figure 4 shows an example of predicted tumor regions by our algorithm compared to previous algorithms that do not consider neighboring patches. We can see that our algorithm introduces very few false positives other than the ground truth tumor regions.
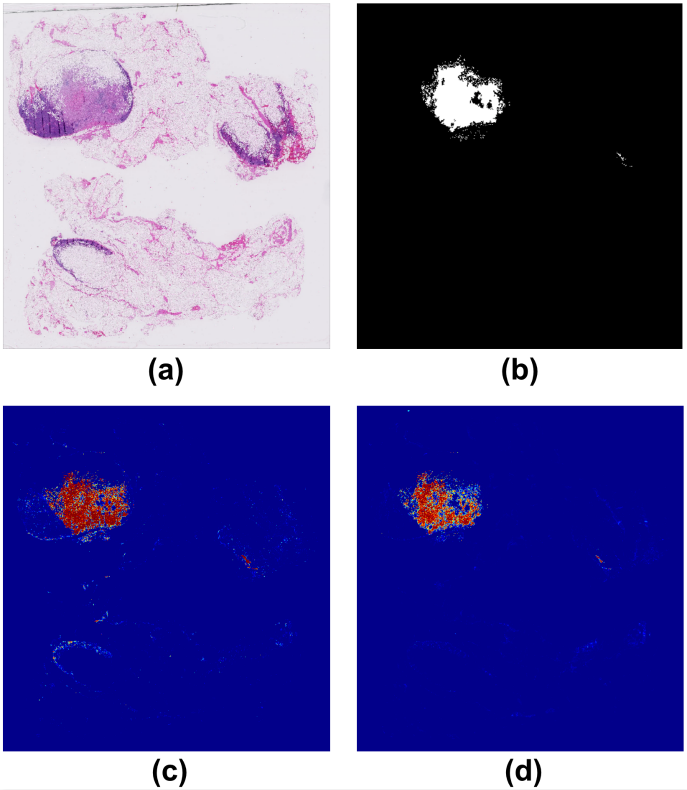
Figure 4. (a) the original whole slide image; (b) annotation by pathologists, where the white regions represent cancer metastasis; (c) predicted tumor regions by previous algorithms that do not consider neighboring patches; (d) predicted tumor regions by our algorithm.
On the test set of the Camelyon16 challenge, our algorithm achieved a tumor localization score (FROC) of 0.8096, which outperforms both a professional pathologist (0.7240), and the previous winner of the Camelyon16 challenge (0.8074). We are also open sourcing our algorithm on Github in the hope to advance the AI research in pathology analysis.
This novel tumor detection algorithm has the potential to improve efficiency and accuracy in pathology slide review. It allows pathologists to focus more on tumor regions highlighted by the algorithm rather than having to search through the whole slide. Further clinical study on a larger dataset is necessary to comprehensively assess the algorithm.
For more details, please refer to our MIDL’ 2018 paper.
1.One pixel is about 0.243µm in digitalized pathology slide at 40X magnification. Micrometastases is often defined as a group of tumor cells with longest diameter larger than 200µm, which is about 823 pixels.